Download this notebook from github.
Workflow Examples
xclim is built on very powerful multiprocessing and distributed computation libraries, notably xarray and dask.
xarray is a python package making it easy to work with n-dimensional arrays. It labels axes with their names [time, lat, lon, level] instead of indices [0,1,2,3], reducing the likelihood of bugs and making the code easier to understand. One of the key strengths of xarray is that it knows how to deal with non-standard calendars (we’re looking at you, “360_days”) and can easily resample daily time series to weekly, monthly, seasonal or annual periods. Finally, xarray is
tightly integrated with dask, a package that can automatically parallelize operations.
The following are a few examples to consult when using xclim to subset NetCDF arrays and compute climate indicators, taking advantage of the parallel processing capabilities offered by xarray and dask. For more information about these projects, please see their documentation pages:
Environment configuration
[1]:
from __future__ import annotations
# File handling libraries
import tempfile
from pathlib import Path
import xarray as xr
# Set xarray to use HTML for displaying outputs
xr.set_options(display_style="html")
import xclim.indices
# Output folder
output_folder = Path(tempfile.mkdtemp())
Setting up the Dask client: parallel processing
In this example, we are using the **dask.distributed** submodule. This is not installed by default in a basic xclim installation. Be sure to add distributed to your Python installation before setting up parallel processing operations!
First, we create a pool of workers that will wait for jobs. The xarray library will automatically connect to these workers and dispatch them jobs that can be run in parallel.
The dashboard link lets you see in real time how busy those workers are.
This step is not mandatory, as dask will fall back to its “single machine scheduler” if a Client is not created. However, this default scheduler doesn’t allow you to set the number of threads or a memory limit and doesn’t start the dashboard, which can be quite useful to understand your task’s progress.
[2]:
from distributed import Client
# Depending on your workstation specifications, you may need to adjust these values.
# On a single machine, n_workers=1 is usually better.
client = Client(n_workers=1, threads_per_worker=4, memory_limit="4GB")
client
[2]:
Client
Client-b2f17bd1-d597-11ee-9875-0242ac110002
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
9b8c4941
| Dashboard: http://127.0.0.1:8787/status | Workers: 1 |
| Total threads: 4 | Total memory: 3.73 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-b69fb0ef-146d-4cbc-bd5b-cca3c58c3e57
| Comm: tcp://127.0.0.1:36047 | Workers: 1 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 4 |
| Started: Just now | Total memory: 3.73 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:42379 | Total threads: 4 |
| Dashboard: http://127.0.0.1:40399/status | Memory: 3.73 GiB |
| Nanny: tcp://127.0.0.1:33409 | |
| Local directory: /tmp/dask-scratch-space/worker-np0r8adv | |
Creating xarray datasets
To open a NetCDF file with xarray, we use xr.open_dataset(<path to file>). By default, the entire file is stored in one chunk, so there is no parallelism. To trigger parallel computations, we need to explicitly specify the chunk size.
In this example, instead of opening a local file, we pass an OPeNDAP URL to xarray. It retrieves the data automatically. Notice also that opening the dataset is quite fast. In fact, the data itself has not been downloaded yet, only the coordinates and the metadata. The downloads will be triggered only when the values need to be accessed directly.
dask’s parallelism is based on memory chunks; We need to tell xarray to split our NetCDF array into chunks of a given size, and operations on each chunk of the array will automatically be dispatched to the workers.
[3]:
data_url = "https://pavics.ouranos.ca/twitcher/ows/proxy/thredds/dodsC/datasets/simulations/bias_adjusted/cmip5/ouranos/cb-oura-1.0/day_ACCESS1-3_historical+rcp85_r1i1p1_na10kgrid_qm-moving-50bins-detrend_1950-2100.ncml"
[4]:
# Chunking in memory along the time dimension.
# Note that the data type is a 'dask.array'. xarray will automatically use client workers.
ds = xr.open_dataset(
data_url,
chunks={"time": 365, "lat": 168, "lon": 150},
drop_variables=["ts", "time_vectors"],
)
print(ds)
<xarray.Dataset> Size: 169GB
Dimensions: (lat: 320, lon: 797, time: 55152)
Coordinates:
* lat (lat) float32 1kB 66.62 66.54 66.46 66.37 ... 40.21 40.12 40.04
* lon (lon) float32 3kB -120.8 -120.7 -120.6 ... -54.63 -54.55 -54.46
* time (time) datetime64[ns] 441kB 1950-01-01 1950-01-02 ... 2100-12-31
Data variables:
tasmin (time, lat, lon) float32 56GB dask.array<chunksize=(365, 168, 150), meta=np.ndarray>
tasmax (time, lat, lon) float32 56GB dask.array<chunksize=(365, 168, 150), meta=np.ndarray>
pr (time, lat, lon) float32 56GB dask.array<chunksize=(365, 168, 150), meta=np.ndarray>
Attributes: (12/26)
Conventions: CF-1.5
title: Ouranos standard ensemble of bias-adjusted cl...
history: CMIP5 compliant file produced from raw ACCESS...
institution: Ouranos Consortium on Regional Climatology an...
source: ACCESS1-3 2011. Atmosphere: AGCM v1.0 (N96 gr...
driving_experiment: historical,rcp85
... ...
frequency: day
modeling_realm: atmos
target_dataset: CANADA : ANUSPLIN interpolated Canada daily 3...
target_dataset_references: CANADA : https://doi.org/10.1175/2011BAMS3132...
driving_institution: Commonwealth Scientific and Industrial Resear...
driving_institute_id: CSIRO-BOM
[5]:
print(ds.tasmin.chunks)
((365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 37), (168, 152), (150, 150, 150, 150, 150, 47))
Multi-file datasets
NetCDF files are often split into periods to keep file size manageable. A single dataset can be split in dozens of individual files. xarray has a function open_mfdataset that can open and aggregate a list of files and construct a unique logical dataset. open_mfdataset can aggregate files over coordinates (time, lat, lon) and variables.
Note that opening a multi-file dataset automatically chunks the array (one chunk per file).
Note also that because
xarrayreads every file metadata to place it in a logical order, it can take a while to load.
[6]:
## Create multi-file data & chunks
# ds = xr.open_mfdataset('/path/to/files*.nc')
Subsetting and selecting data with xarray
Here, we will reduce the size of our data using the methods implemented in xarray (docs here).
[7]:
ds2 = ds.sel(lat=slice(50, 45), lon=slice(-70, -65), time=slice("2090", "2100"))
print(ds2.tasmin)
<xarray.DataArray 'tasmin' (time: 4017, lat: 60, lon: 60)> Size: 58MB
dask.array<getitem, shape=(4017, 60, 60), dtype=float32, chunksize=(365, 60, 60), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 240B 49.96 49.87 49.79 49.71 ... 45.21 45.12 45.04
* lon (lon) float32 240B -69.96 -69.88 -69.8 ... -65.21 -65.13 -65.05
* time (time) datetime64[ns] 32kB 2090-01-01 2090-01-02 ... 2100-12-31
Attributes:
long_name: air_temperature
standard_name: air_temperature
units: K
_ChunkSizes: [256 16 16]
[8]:
ds3 = ds.sel(lat=46.8, lon=-71.22, method="nearest").sel(time="1993")
print(ds3.tasmin)
<xarray.DataArray 'tasmin' (time: 365)> Size: 1kB
dask.array<getitem, shape=(365,), dtype=float32, chunksize=(354,), chunktype=numpy.ndarray>
Coordinates:
lat float32 4B 46.79
lon float32 4B -71.21
* time (time) datetime64[ns] 3kB 1993-01-01 1993-01-02 ... 1993-12-31
Attributes:
long_name: air_temperature
standard_name: air_temperature
units: K
_ChunkSizes: [256 16 16]
For more powerful subsetting tools with features such as coordinate reference system (CRS) aware subsetting and vector shape masking, the xclim developers strongly encourage users to consider the subsetting utilities of the clisops package.
Their documentation showcases several examples of how to perform more complex subsetting: clisops.core.subset.
Climate index calculation & resampling frequencies
xclim has two layers for the calculation of indicators. The bottom layer is composed of a list of functions that take one or more xarray.DataArray’s as input and return an xarray.DataArray as output. You’ll find these functions in xclim.indices. The indicator’s logic is contained in this function, as well as some unit handling, but it doesn’t perform any data consistency checks (like if the time frequency is daily), and doesn’t adjust the metadata of the output array.
The second layer are class instances that you’ll find organized by realm. So far, there are three realms available in xclim.atmos, xclim.seaIce and xclim.land, the first one being the most exhaustive. Before running computations, these classes check if the input data is a daily average of the expected variable:
If an indicator expects a daily mean, and you pass it a daily max, a
warningwill be raised.After the computation, it also checks the number of values per period to make sure there are not missing values or
NaNin the input data. If there are, the output is going to be set toNaN. Ex. : If the indicator performs a yearly resampling, but there are only 350 non-NaNvalues in one given year in the input data, that year’s output will beNaN.The output units are set correctly as well as other properties of the output array, complying as much as possible with CF conventions.
For new users, we suggest you use the classes found in xclim.atmos and others. If you know what you’re doing, and you want to circumvent the built-in checks, then you can use the xclim.indices directly.
Almost all xclim indicators convert daily data to lower time frequencies, such as seasonal or annual values. This is done using xarray.DataArray.resample method. Resampling creates a grouped object over which you apply a reduction operation (e.g. mean, min, max). The list of available frequency is given in the link below, but the most often used are:
YS: annual starting in JanuaryYS-JUL: annual starting in JulyMS: monthlyQS-DEC: seasonal starting in December
More info about this specification can be found in pandas’ documentation
Note - not all offsets in the link are supported by cftime objects in xarray.
In the example below, we’re computing the annual maximum temperature of the daily maximum temperature (tx_max).
[9]:
out = xclim.atmos.tx_max(ds2.tasmax, freq="YS")
print(out)
<xarray.DataArray 'tx_max' (time: 11, lat: 60, lon: 60)> Size: 158kB
dask.array<where, shape=(11, 60, 60), dtype=float32, chunksize=(1, 60, 60), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 240B 49.96 49.87 49.79 49.71 ... 45.21 45.12 45.04
* lon (lon) float32 240B -69.96 -69.88 -69.8 ... -65.21 -65.13 -65.05
* time (time) datetime64[ns] 88B 2090-01-01 2091-01-01 ... 2100-01-01
Attributes:
long_name: Maximum daily maximum temperature
standard_name: air_temperature
units: K
_ChunkSizes: [256 16 16]
cell_methods: time: maximum over days
history: [2024-02-27 17:43:23] tx_max: TX_MAX(tasmax=tasmax, freq=...
description: Annual maximum of daily maximum temperature.
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
If you execute the cell above, you’ll see that this operation is quite fast. This a feature coming from dask. Read Lazy computation further down.
Comparison of atmos vs indices modules
Using the xclim.indices module performs not checks and only fills the units attribute.
[10]:
out = xclim.indices.tx_days_above(ds2.tasmax, thresh="30 degC", freq="YS")
print(out)
<xarray.DataArray 'tasmax' (time: 11, lat: 60, lon: 60)> Size: 317kB
dask.array<mul, shape=(11, 60, 60), dtype=float64, chunksize=(1, 60, 60), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 240B 49.96 49.87 49.79 49.71 ... 45.21 45.12 45.04
* lon (lon) float32 240B -69.96 -69.88 -69.8 ... -65.21 -65.13 -65.05
* time (time) datetime64[ns] 88B 2090-01-01 2091-01-01 ... 2100-01-01
Attributes:
units: d
With xclim.atmos, checks are performed and many CF-compliant attributes are added:
[11]:
out = xclim.atmos.tx_days_above(ds2.tasmax, thresh="30 degC", freq="YS")
print(out)
<xarray.DataArray 'tx_days_above' (time: 11, lat: 60, lon: 60)> Size: 317kB
dask.array<where, shape=(11, 60, 60), dtype=float64, chunksize=(1, 60, 60), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 240B 49.96 49.87 49.79 49.71 ... 45.21 45.12 45.04
* lon (lon) float32 240B -69.96 -69.88 -69.8 ... -65.21 -65.13 -65.05
* time (time) datetime64[ns] 88B 2090-01-01 2091-01-01 ... 2100-01-01
Attributes:
units: days
cell_methods: time: sum over days
history: [2024-02-27 17:43:24] tx_days_above: TX_DAYS_ABOVE(tasmax...
standard_name: number_of_days_with_air_temperature_above_threshold
long_name: The number of days with maximum temperature above 30 degc
description: Annual number of days where daily maximum temperature exc...
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
[12]:
# We have created an xarray data-array - We can insert this into an output xr.Dataset object with a copy of the original dataset global attrs
ds_out = xr.Dataset(attrs=ds2.attrs)
# Add our climate index as a data variable to the dataset
ds_out[out.name] = out
print(ds_out)
<xarray.Dataset> Size: 317kB
Dimensions: (lat: 60, lon: 60, time: 11)
Coordinates:
* lat (lat) float32 240B 49.96 49.87 49.79 ... 45.21 45.12 45.04
* lon (lon) float32 240B -69.96 -69.88 -69.8 ... -65.13 -65.05
* time (time) datetime64[ns] 88B 2090-01-01 ... 2100-01-01
Data variables:
tx_days_above (time, lat, lon) float64 317kB dask.array<chunksize=(1, 60, 60), meta=np.ndarray>
Attributes: (12/26)
Conventions: CF-1.5
title: Ouranos standard ensemble of bias-adjusted cl...
history: CMIP5 compliant file produced from raw ACCESS...
institution: Ouranos Consortium on Regional Climatology an...
source: ACCESS1-3 2011. Atmosphere: AGCM v1.0 (N96 gr...
driving_experiment: historical,rcp85
... ...
frequency: day
modeling_realm: atmos
target_dataset: CANADA : ANUSPLIN interpolated Canada daily 3...
target_dataset_references: CANADA : https://doi.org/10.1175/2011BAMS3132...
driving_institution: Commonwealth Scientific and Industrial Resear...
driving_institute_id: CSIRO-BOM
Different ways of resampling
Many indices use algorithms that find the length of given sequences. For instance, xclim.indices.heat_wave_max_length finds the longest sequence where tasmax and tasmin are above given threshold values. Resampling can be used to find the longest sequence in given periods of time, for instance the longest heat wave for each month if the resampling frequency is freq == "MS".
The order of the two operations just described, i.e. : * Finding the length of sequences respecting a certain criterion (“run length algorithms”) * Separating the dataset in given time periods (“resampling”)
is important and can lead to differing results.
The cell below illustrates this by looking at the maximum lengths of heat waves each month from May 2010 to August 2010 by doing these operations in the two possible orders. The heat wave max lengths for July in a small region of interest \(\text{lat} \in [43, 44.5],\, \text{lon} \in [-117.5, -116]\) are shown: The maximal lengths are sometimes longer first applying the run length algorithm (resample_before_rl == False).
[13]:
# import plotting stuff
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams["figure.figsize"] = (11, 5)
[14]:
ds_reduced = (
ds.sel(lat=slice(44.5, 43))
.sel(lon=slice(-117.5, -116))
.sel(time=slice("2010-05-01", "2011-08-31"))
)
tn, tx = ds_reduced.tasmin, ds_reduced.tasmax
freq = "MS"
# Threshold on tasmin: chosen smaller than the default 22.0 degC
thresh_tn = "20.0 degC"
# Computing index by resampling **before** run length algorithm (default value)
hw_before = xclim.indices.heat_wave_max_length(
tn, tx, freq=freq, thresh_tasmin=thresh_tn, resample_before_rl=True
)
# Computing index by resampling **after** run length algorithm
hw_after = xclim.indices.heat_wave_max_length(
tn, tx, freq=freq, thresh_tasmin=thresh_tn, resample_before_rl=False
)
hw_before.sel(time="2010-07-01").plot(vmin=0, vmax=7)
plt.title("Resample, then run length")
plt.figure()
hw_after.sel(time="2010-07-01").plot(vmin=0, vmax=7)
plt.title("Run length, then resample")
plt.show()
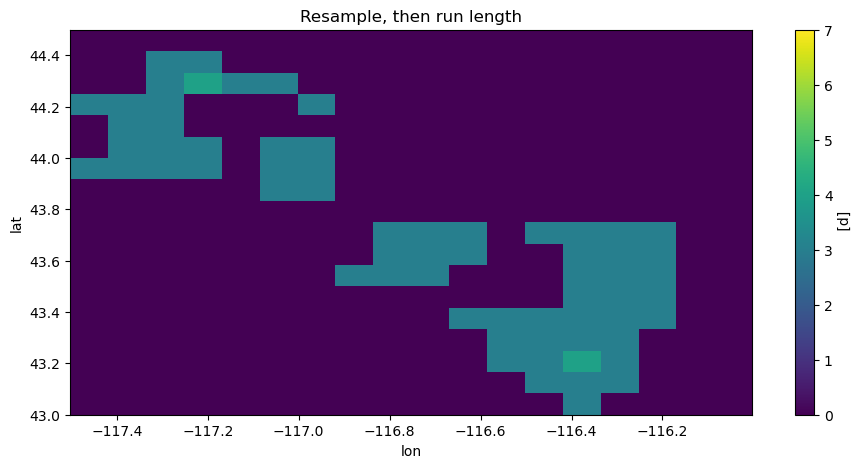
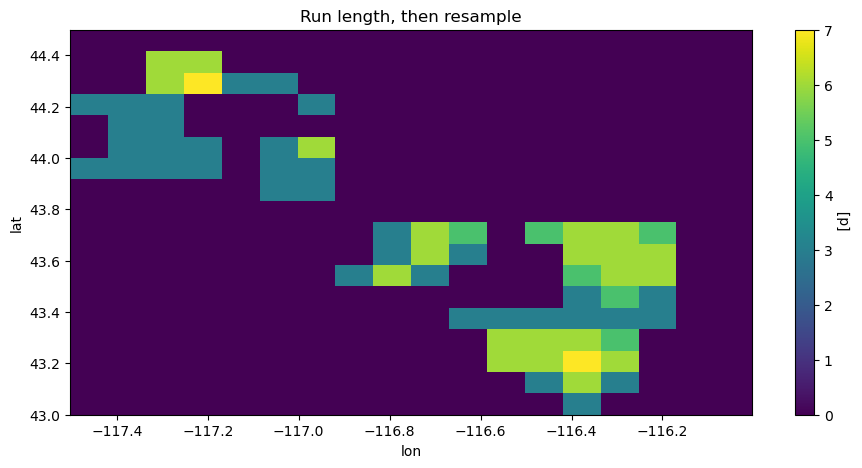
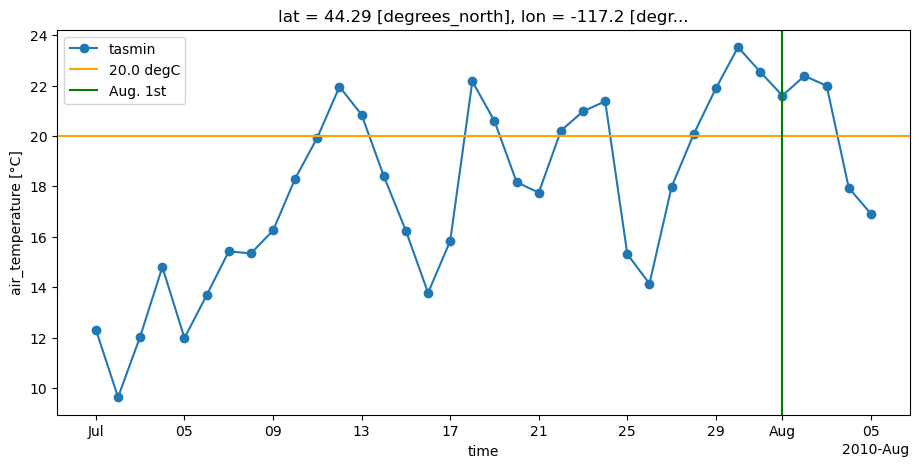
Let’s focus on the point \((-117.21075, 44.29087)\), which has a maximum wave length of four (4) or seven (7), depending on whether resampling occurs before or after the run length algorithm.
Plotting the values of tasmin in July and early August, we see a sequence of seven hot minimal temperatures at the end of July that surpass the threshold to qualify for a heat wave.
If resampling occurs first, and we first separate the periods in months, the run length algorithms will only look for sequences of hot days within the month of July and will exclude the last 3 days of this sequence of 7 days.
Using the run length algorithm before resampling looks for sequences of hot days in all the dataset given (temperatures from May 1, 2010 to Aug. 31, 2010) and then subdivides these sequences in the months where they have started. Since it starts in July, this sequence is registered as counts for a heat wave of seven days happening in July.
This also implies that the first 3 days of August which belong in this sequence of seven days will be counted as a sequence in August with the first method, but not with the second.
[15]:
from xclim.core.units import convert_units_to
# Select a spatial point of interest in July-early August
lon_i, lat_i = -117.21075, 44.29087
tn_pt = tn.sel(time=slice("2010-07-01", "2010-08-05")).sel(lat=lat_i, lon=lon_i)
tn_pt = convert_units_to(tn_pt, "degC")
tn_pt.plot(marker="o", label="tasmin")
plt.axhline(y=convert_units_to(thresh_tn, "degC"), color="orange", label=thresh_tn)
plt.axvline(x=["2010-08-01"], color="green", label="Aug. 1st")
plt.legend()
plt.show()
Lazy computation - Nothing has been computed so far !
If you look at the output of those operations, they’re identified as dask.array objects. What happens is that dask creates a chain of operations that, when executed, will yield the values we want. We have thus far only created a schedule of tasks with a small preview and not done any actual computations. You can trigger computations by using the load or compute method, or writing the output to disk via to_netcdf. Of course, calling .plot() will also trigger the
computation.
[16]:
%%time
output_file = output_folder / "test_tx_max.nc"
dsOut.to_netcdf(output_file)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
File <timed exec>:2
NameError: name 'dsOut' is not defined
(Times may of course vary depending on the machine and the Client settings)
Performance tips
Optimizing the chunk size
You can improve performance by being smart about chunk sizes. If chunks are too small, there is a lot of time lost in overhead. If chunks are too large, you may end up exceeding the individual worker memory limit.
[17]:
print(ds2.chunks["time"])
(330, 365, 365, 365, 365, 365, 365, 365, 365, 365, 365, 37)
[18]:
# rechunk data in memory for the entire grid
ds2c = ds2.chunk(chunks={"time": 4 * 365})
print(ds2c.chunks["time"])
(1460, 1460, 1097)
[19]:
%%time
out = xclim.atmos.tx_max(ds2c.tasmax, freq="YS")
ds_out = xr.Dataset(data_vars=None, coords=out.coords, attrs=ds.attrs)
ds_out[out.name] = out
output_file = output_folder / "test_tx_max.nc"
ds_out.to_netcdf(output_file)
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
CPU times: user 351 ms, sys: 39.3 ms, total: 391 ms
Wall time: 11.1 s
Loading the data in memory
If the dataset is relatively small, it might be more efficient to simply load the data into the memory and use numpy arrays instead of dask arrays.
[20]:
ds4 = ds3.load()
Unit handling in xclim
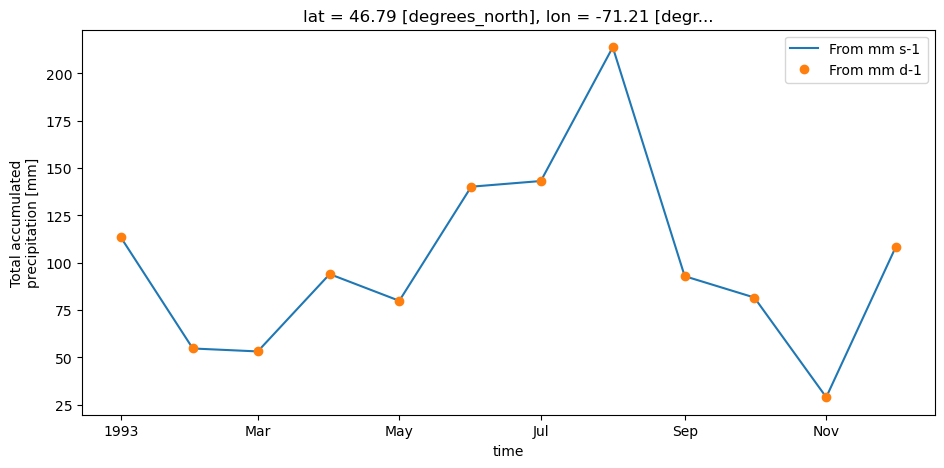
A lot of effort has been placed into automatic handling of input data units. xclim will automatically detect the input variable(s) units (e.g. °C versus °K or mm/s versus mm/day etc.) and adjust on-the-fly in order to calculate indices in the consistent manner. This comes with the obvious caveat that input data requires metadata attribute for units.
The Units Handling page goes more into detail on how unit conversion can easily be done.
In the example below, we compute weekly total precipitation in mm using inputs of mm/s and mm/d. As we can see, the output is identical.
[21]:
# Compute with the original mm s-1 data
out1 = xclim.atmos.precip_accumulation(ds4.pr, freq="MS")
# Create a copy of the data converted to mm d-1
pr_mmd = ds4.pr * 3600 * 24
pr_mmd.attrs["units"] = "mm d-1"
out2 = xclim.atmos.precip_accumulation(pr_mmd, freq="MS")
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:46: UserWarning: Variable has a non-conforming standard_name: Got `lwe_precipitation_rate`, expected `['precipitation_flux']`
check_valid(vardata, "standard_name", data["standard_name"])
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:46: UserWarning: Variable does not have a `standard_name` attribute.
check_valid(vardata, "standard_name", data["standard_name"])
[22]:
plt.figure()
out1.plot(label="From mm s-1", linestyle="-")
out2.plot(label="From mm d-1", linestyle="none", marker="o")
plt.legend()
plt.show()
Threshold indices
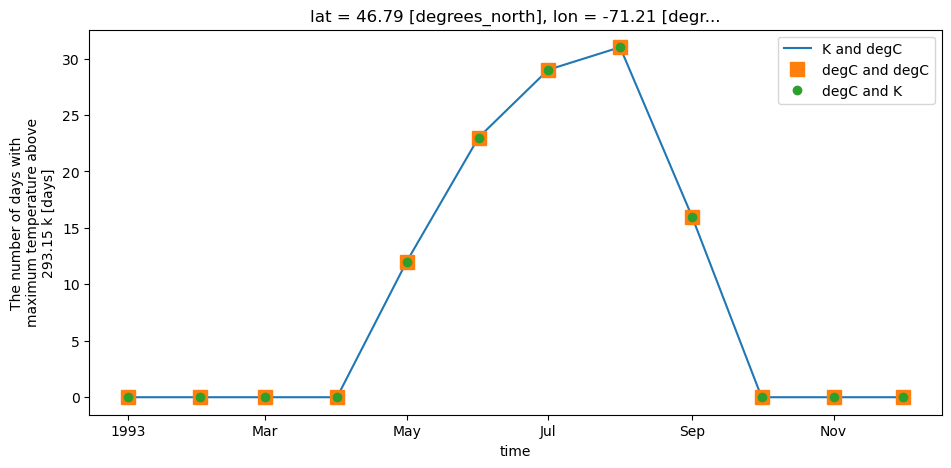
xclim unit handling also applies to threshold indicators. Users can provide threshold in units of choice and xclim will adjust automatically. For example, determining the number of days with tasmax > 20 °C, users can define a threshold input of "20 C" or "20 degC" even if input data is in Kelvin. Alternatively, users can even provide a threshold in Kelvin ("293.15 K", if they really wanted to).
[23]:
# Create a copy of the data converted to C
tasmax_C = ds4.tasmax - 273.15
tasmax_C.attrs["units"] = "C"
# Using Kelvin data, threshold in Celsius
out1 = xclim.atmos.tx_days_above(ds4.tasmax, thresh="20 C", freq="MS")
# Using Celsius data
out2 = xclim.atmos.tx_days_above(tasmax_C, thresh="20 C", freq="MS")
# Using Celsius but with threshold in Kelvin
out3 = xclim.atmos.tx_days_above(tasmax_C, thresh="293.15 K", freq="MS")
# Plot and see that it's all identical:
plt.figure()
out1.plot(label="K and degC", linestyle="-")
out2.plot(label="degC and degC", marker="s", markersize=10, linestyle="none")
out3.plot(label="degC and K", marker="o", linestyle="none")
plt.legend()
plt.show()
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:46: UserWarning: Variable does not have a `standard_name` attribute.
check_valid(vardata, "standard_name", data["standard_name"])
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:46: UserWarning: Variable does not have a `standard_name` attribute.
check_valid(vardata, "standard_name", data["standard_name"])
Spatially varying thresholds
Thresholds can also be passed as DataArrays instead of single scalar values, allowing the computation to depend on one or more non-temporal dimensions. The units attribute must be set.
Going back to the initial ds, we’ll subset it and compute the length of the heat wave according to thresholds that vary along the latitude and longitude.
[24]:
ds5 = ds.sel(time=slice("1950", "1960"), lat=slice(50, 46), lon=slice(-75, -71))
ds5
[24]:
<xarray.Dataset> Size: 111MB
Dimensions: (lat: 48, lon: 48, time: 4018)
Coordinates:
* lat (lat) float32 192B 49.96 49.87 49.79 49.71 ... 46.21 46.12 46.04
* lon (lon) float32 192B -74.96 -74.88 -74.8 ... -71.21 -71.13 -71.05
* time (time) datetime64[ns] 32kB 1950-01-01 1950-01-02 ... 1960-12-31
Data variables:
tasmin (time, lat, lon) float32 37MB dask.array<chunksize=(365, 48, 48), meta=np.ndarray>
tasmax (time, lat, lon) float32 37MB dask.array<chunksize=(365, 48, 48), meta=np.ndarray>
pr (time, lat, lon) float32 37MB dask.array<chunksize=(365, 48, 48), meta=np.ndarray>
Attributes: (12/26)
Conventions: CF-1.5
title: Ouranos standard ensemble of bias-adjusted cl...
history: CMIP5 compliant file produced from raw ACCESS...
institution: Ouranos Consortium on Regional Climatology an...
source: ACCESS1-3 2011. Atmosphere: AGCM v1.0 (N96 gr...
driving_experiment: historical,rcp85
... ...
frequency: day
modeling_realm: atmos
target_dataset: CANADA : ANUSPLIN interpolated Canada daily 3...
target_dataset_references: CANADA : https://doi.org/10.1175/2011BAMS3132...
driving_institution: Commonwealth Scientific and Industrial Resear...
driving_institute_id: CSIRO-BOM[25]:
# The tasmin threshold is 7°C for the northern half of the domain and 11°C for the southern half.
# (notice that the lat coordinate is in decreasing order : from north to south)
thresh_tasmin = xr.DataArray(
[7] * 24 + [11] * 24, dims=("lat",), coords={"lat": ds5.lat}, attrs={"units": "°C"}
)
# The tasmax threshold is 17°C for the western half of the domain and 21°C for the eastern half.
thresh_tasmax = xr.DataArray(
[17] * 24 + [21] * 24, dims=("lon",), coords={"lon": ds5.lon}, attrs={"units": "°C"}
)
out_hw2d = xclim.atmos.heat_wave_total_length(
tasmin=ds5.tasmin,
tasmax=ds5.tasmax,
thresh_tasmin=thresh_tasmin,
thresh_tasmax=thresh_tasmax,
freq="YS",
window=3,
)
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/stable/lib/python3.12/site-packages/xclim/core/cfchecks.py:42: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
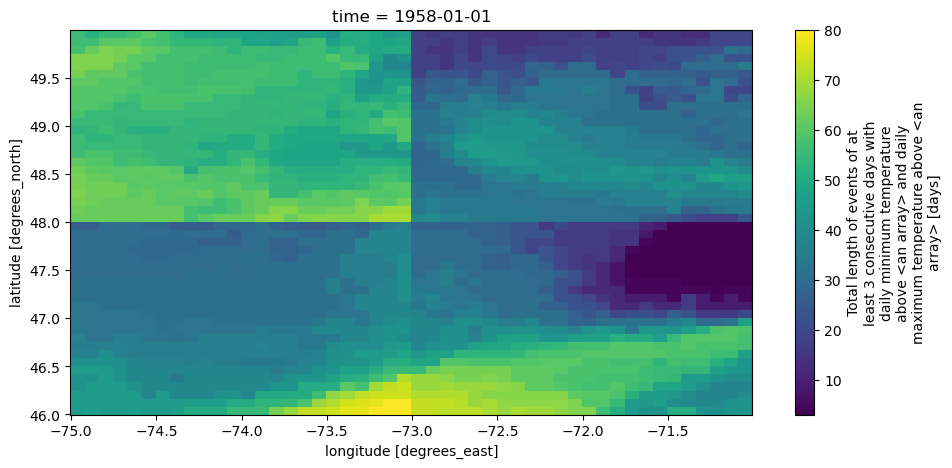
The final map for year 1958, shows clear jumps across the 4 quadrants, which was expected with our space-dependent thresholds. Notice also how the long_name (printed on the colorbar label) mentions that the threshold comes from “an array”. This imprecise metadata is a consequence of using DataArray-derived thresholds.
[26]:
out_hw2d.sel(time="1958").plot()
plt.show()