Download this notebook from github.
Basic Usage
Climate indicator computations
xclim is a library of climate indicators that operate on xarray DataArray objects.
xclim provides two layers of computations, one responsible for computations and units handling (the computation layer, the indices), and the other responsible for input health checks and metadata formatting (the CF layer, referring to the Climate and Forecast convention, the indicators). Functions from the computation layer are found in xclim.indices, while indicator objects from the CF layer are found in realm modules (xclim.atmos, xclim.land and xclim.seaIce).
Users should always use the indicators, and maybe revert to indices as a last resort if the indicator machinery becomes too heavy for their special edge case.
To use xclim in a project, import both xclim and xarray.
[1]:
from __future__ import annotations
import xarray as xr
import xclim
from xclim import testing
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/indices/fire/_cffwis.py:207: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
def _day_length(lat: int | float, mth: int): # pragma: no cover
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/indices/fire/_cffwis.py:227: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
def _day_length_factor(lat: float, mth: int): # pragma: no cover
Index calculations are performed by opening a NetCDF-like file, accessing the variable of interest, and calling the index function, which returns a new xarray.DataArray.
For this example, we’ll first open a demonstration dataset storing surface air temperature and compute the number of growing degree days (the sum of degrees above a certain threshold) at the monthly frequency.
[2]:
# Normally, we would use xarray to open a dataset, e.g.:
# ds = xr.open_dataset("your_file.nc")
# For this example, let's use a test dataset from xclim:
ds = testing.open_dataset("ERA5/daily_surface_cancities_1990-1993.nc")
ds.tas
[2]:
<xarray.DataArray 'tas' (location: 5, time: 1461)>
[7305 values with dtype=float32]
Coordinates:
lon (location) float32 ...
* location (location) object 'Halifax' 'Montréal' ... 'Saskatoon' 'Victoria'
lat (location) float32 ...
* time (time) datetime64[ns] 1990-01-01 1990-01-02 ... 1993-12-31
Attributes:
cell_methods: time: mean within days
long_name: Mean daily surface temperature
original_variable: t2m
standard_name: air_temperature
units: K[3]:
gdd = xclim.atmos.growing_degree_days(tas=ds.tas, thresh="10.0 degC", freq="YS")
gdd
[3]:
<xarray.DataArray 'growing_degree_days' (location: 5, time: 4)>
array([[7.76921753e+02, 7.03898560e+02, 6.31244263e+02, 6.51882751e+02],
[1.22686438e+03, 1.35817456e+03, 1.12669324e+03, 1.22071863e+03],
[6.99319458e+00, 2.39197693e+01, 6.53686523e-01, 1.87173157e+01],
[9.29841431e+02, 1.00834296e+03, 7.17684204e+02, 6.29096436e+02],
[5.87043213e+02, 5.02022339e+02, 5.99313477e+02, 5.67815552e+02]],
dtype=float32)
Coordinates:
lon (location) float32 -63.4 -73.4 -68.4 -106.7 -123.2
* location (location) object 'Halifax' 'Montréal' ... 'Saskatoon' 'Victoria'
lat (location) float32 44.5 45.5 63.75 52.0 48.5
* time (time) datetime64[ns] 1990-01-01 1991-01-01 1992-01-01 1993-01-01
Attributes:
units: K days
cell_methods: time: mean within days time: sum over days
history: [2023-05-09 21:53:17] growing_degree_days: GROWING_DEGREE...
standard_name: integral_of_air_temperature_excess_wrt_time
long_name: Cumulative sum of temperature degrees for mean daily temp...
description: Annual growing degree days (mean temperature above 10.0 d...This computation was made using the growing_degree_days indicator. The same computation could be made through the index. You can see how the metadata is alot poorer here.
[4]:
gdd = xclim.indices.growing_degree_days(tas=ds.tas, thresh="10.0 degC", freq="YS")
gdd
[4]:
<xarray.DataArray 'tas' (location: 5, time: 4)>
array([[7.76921753e+02, 7.03898560e+02, 6.31244263e+02, 6.51882751e+02],
[1.22686438e+03, 1.35817456e+03, 1.12669324e+03, 1.22071863e+03],
[6.99319458e+00, 2.39197693e+01, 6.53686523e-01, 1.87173157e+01],
[9.29841431e+02, 1.00834296e+03, 7.17684204e+02, 6.29096436e+02],
[5.87043213e+02, 5.02022339e+02, 5.99313477e+02, 5.67815552e+02]],
dtype=float32)
Coordinates:
lon (location) float32 -63.4 -73.4 -68.4 -106.7 -123.2
* location (location) object 'Halifax' 'Montréal' ... 'Saskatoon' 'Victoria'
lat (location) float32 44.5 45.5 63.75 52.0 48.5
* time (time) datetime64[ns] 1990-01-01 1991-01-01 1992-01-01 1993-01-01
Attributes:
units: K dThe call to xclim.indices.growing_degree_days first checked that the input variable units were units of temperature, ran the computation, then set the output’s units to the appropriate unit (here "K d" or Kelvin days). As you can see, the Indicator returned the same output, but with more metadata, it also performed more checks as explained below.
growing_degree_days makes most sense with daily input, but could theoretically accept other source frequencies. The computational layer (``Index``) assumes that users have checked that the input data has the expected temporal frequency and has no missing values. However, no checks are performed, so the output data could be wrong (which is why it’s always safer to use ``Indicator`` objects from the CF layer, as demonstrated in the following section).
Finally, as almost all indices, the function takes a freq argument to specify over what time period it is computed. These are called “Offset Aliases” and are the same as the resampling string arguments. Valid arguments are detailed in pandas docs (note that aliases involving “business” notions are not supported by xarray and thus could raise issues in xclim.
Units Handling paradigm
Indices are written in order to be flexible as to the sampling frequency and units of the data. You can use growing_degree_days on, for example, the 6-hourly data, but the output will then be in degree-hour units ("K h"). Moreover, all units, even when untouched by the calculation, will be reformatted into a CF-compliant symbol format. This behaviour was chosen to ensure consistency between all indices.
Very few indices will convert their output to specific units; Rather, it is the dimensionality that will be consistent on output. The Units Handling page goes more into detail on how unit conversion can easily be done.
This doesn’t apply to ``Indicators``. Those will always output data in a specific unit, the one listed in the Indicators.cf_attrs metadata dictionary.
Conventions
As you may have noticed, the growing_degree_days function above was not told along which dimension to operate. In xclim, the temporal dimension is always assumed to be named "time". All functions which reduce or compute over that dimension will expect that name. If you ever have another name in your data, you can simply rename it like:
ds = ds.rename(T="time")
For other names and attributes, xclim tries to follow different sets of conventions. In particular, input data should follow the CF conventions whenever possible for variable attributes. Variable names are usually the ones used in CMIP6, when they exist.
Indicators
Indices with Health Checks and Metadata Attributes
Indicator instances from the CF layer are found in modules bearing the name of the computational realm in which its input variables are typically found: xclim.atmos, xclim.land and xclim.seaIce. These objects run sanity checks on the input variables and set output’s metadata according to CF-conventions when applicable. Some checks involve:
Identifying periods where missing data significantly impacts the calculation and omits calculations for those periods. Those are called “missing methods” and are detailed in section Health checks.
Appending process history and maintaining the historical provenance of file metadata.
Writing Climate and Forecast Convention compliant metadata based on the variables and indices calculated.
Those modules are best used for producing NetCDF files that will be shared with users. See Climate Indicators for a list of available indicators.
If we run the growing_degree_days indicator over a non-daily dataset, we’ll be warned that the input data is not daily. That is, running xclim.atmos.growing_degree_days(ds.air, thresh='10.0 degC', freq='MS') will fail with a ValidationError:
[5]:
# Show that data is not at a daily time frequency
ds6h = xr.tutorial.open_dataset("air_temperature")
xr.infer_freq(ds6h.time)
[5]:
'6H'
[6]:
gdd = xclim.atmos.growing_degree_days(tas=ds6h.air, thresh="10.0 degC", freq="MS")
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
ValueError: Frequency of time series not strictly in ['D']. To mute this, set xclim's option data_validation='log'.
The above exception was the direct cause of the following exception:
ValidationError Traceback (most recent call last)
Cell In[6], line 1
----> 1 gdd = xclim.atmos.growing_degree_days(tas=ds6h.air, thresh="10.0 degC", freq="MS")
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:832, in Indicator.__call__(self, *args, **kwds)
829 out_attrs = {}
830 out_attrs = [out_attrs.copy() for i in range(self.n_outs)]
--> 832 das, params = self._preprocess_and_checks(das, params)
834 # Get correct variable names for the compute function.
835 inv_var_map = dict(map(reversed, self._variable_mapping.items()))
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:1489, in ResamplingIndicatorWithIndexing._preprocess_and_checks(self, das, params)
1487 def _preprocess_and_checks(self, das: dict[str, DataArray], params: dict[str, Any]):
1488 """Perform parent's checks and also check if freq is allowed."""
-> 1489 das, params = super()._preprocess_and_checks(das, params)
1491 indxr = params.get("indexer")
1492 if indxr:
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:1409, in ResamplingIndicator._preprocess_and_checks(self, das, params)
1407 def _preprocess_and_checks(self, das, params):
1408 """Perform parent's checks and also check if freq is allowed."""
-> 1409 das, params = super()._preprocess_and_checks(das, params)
1411 # Check if the period is allowed:
1412 if (
1413 self.allowed_periods is not None
1414 and parse_offset(params["freq"])[1] not in self.allowed_periods
1415 ):
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:947, in Indicator._preprocess_and_checks(self, das, params)
945 """Actions to be done after parsing the arguments and before computing."""
946 # Pre-computation validation checks on DataArray arguments
--> 947 self._bind_call(self.datacheck, **das)
948 self._bind_call(self.cfcheck, **das)
949 return das, params
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:984, in Indicator._bind_call(self, func, **das)
981 return func(*das.values())
982 else:
983 # Call the func using bound arguments
--> 984 return func(*ba.args, **ba.kwargs)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/indicator.py:1303, in Indicator.datacheck(self, **das)
1301 for key, da in das.items():
1302 if "time" in da.coords and da.time.ndim == 1 and len(da.time) > 3:
-> 1303 datachecks.check_freq(da, self.src_freq, strict=True)
1305 datachecks.check_common_time(
1306 [
1307 da
(...)
1310 ]
1311 )
File <boltons.funcutils.FunctionBuilder-163>:2, in check_freq(var, freq, strict)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/options.py:122, in datacheck.<locals>.run_check(*args, **kwargs)
120 @wraps(func)
121 def run_check(*args, **kwargs):
--> 122 return _run_check(func, DATA_VALIDATION, *args, **kwargs)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/options.py:114, in _run_check(func, option, *args, **kwargs)
112 func(*args, **kwargs)
113 except ValidationError as err:
--> 114 raise_warn_or_log(err, OPTIONS[option], stacklevel=4)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/utils.py:498, in raise_warn_or_log(err, mode, msg, err_type, stacklevel)
496 warnings.warn(msg, stacklevel=stacklevel + 1)
497 else: # mode == "raise"
--> 498 raise err from err_type(msg)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/options.py:112, in _run_check(func, option, *args, **kwargs)
110 """Run function and customize exception handling based on option."""
111 try:
--> 112 func(*args, **kwargs)
113 except ValidationError as err:
114 raise_warn_or_log(err, OPTIONS[option], stacklevel=4)
File ~/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/datachecks.py:53, in check_freq(var, freq, strict)
49 v_base = parse_offset(v_freq)[1]
50 if v_base not in exp_base or (
51 strict and all(compare_offsets(v_freq, "!=", frq) for frq in freq)
52 ):
---> 53 raise ValidationError(
54 f"Frequency of time series not {'strictly' if strict else ''} in {freq}. "
55 "To mute this, set xclim's option data_validation='log'."
56 )
ValidationError: Frequency of time series not strictly in ['D']. To mute this, set xclim's option data_validation='log'.
Resampling to a daily frequency and running the same indicator succeeds, but we will still get warnings from the CF metadata checks.
[7]:
daily_ds = ds6h.resample(time="D").mean(keep_attrs=True)
gdd = xclim.atmos.growing_degree_days(daily_ds.air, thresh="10.0 degC", freq="YS")
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/cfchecks.py:41: UserWarning: Variable does not have a `cell_methods` attribute.
_check_cell_methods(
/home/docs/checkouts/readthedocs.org/user_builds/xclim/conda/v0.43.0/lib/python3.10/site-packages/xclim/core/cfchecks.py:45: UserWarning: Variable does not have a `standard_name` attribute.
check_valid(vardata, "standard_name", data["standard_name"])
To suppress the CF validation warnings, we can set xclim to send these warnings to the log instead of raising a warning or an error. We also could set data_validation='warn' to be able to run the indicator on non-daily data. These options can be set globally or within a context manager with set_options.
The missing method which determines if a period should be considered missing or not can be controlled through the check_missing option, globally or contextually. The main missing methods also have options that can be modified.
[8]:
with xclim.set_options(
check_missing="pct",
missing_options={"pct": dict(tolerance=0.1)},
cf_compliance="log",
):
# Change the missing method to "percent", instead of the default "any"
# Set the tolerance to 10%, periods with more than 10% of missing data
# in the input will be masked in the ouput.
gdd = xclim.atmos.growing_degree_days(daily_ds.air, thresh="10.0 degC", freq="MS")
Some indicators also expose time-selection arguments as **indexer keywords. This allows to run the index on a subset of the time coordinates, for example only on a specific season, month, or between two dates in every year. It relies on the select_time function. Some indicators will simply select the time period and run the calculations, while others will smartly perform the selection at the right time, when the order of operation makes
a difference. All will pass the indexer kwargs to the missing value handling, ensuring that the missing values outside the valid time period are not considered.
The next example computes the annual sum of growing degree days over 10 °C, but only considering days from the 1st of April to the 30th of September.
[9]:
with xclim.set_options(cf_compliance="log"):
gdd = xclim.atmos.growing_degree_days(
tas=daily_ds.air, thresh="10 degC", freq="YS", date_bounds=("04-01", "09-30")
)
gdd
[9]:
<xarray.DataArray 'growing_degree_days' (time: 2, lat: 25, lon: 53)>
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
[0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
[3.3140106e+01, 5.0820068e+01, 6.6547638e+01, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
...,
[2.7736931e+03, 2.6248132e+03, 2.5183257e+03, ...,
2.6201809e+03, 2.5202234e+03, 2.4362007e+03],
[2.8073433e+03, 2.7539409e+03, 2.6544856e+03, ...,
2.6141133e+03, 2.6077134e+03, 2.5585957e+03],
[2.8185557e+03, 2.8164485e+03, 2.7658508e+03, ...,
2.6862107e+03, 2.6818708e+03, 2.6931655e+03]],
[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
[0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
[1.0225220e+00, 5.5400085e+00, 1.0475037e+01, ...,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
...,
[2.8183232e+03, 2.6905308e+03, 2.6107834e+03, ...,
2.5506509e+03, 2.4474634e+03, 2.3652031e+03],
[2.8695334e+03, 2.8242583e+03, 2.7269106e+03, ...,
2.5259932e+03, 2.5199482e+03, 2.4677581e+03],
[2.8881082e+03, 2.8856885e+03, 2.8283706e+03, ...,
2.5869858e+03, 2.5948560e+03, 2.6111182e+03]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 2014-01-01
Attributes:
units: K days
cell_methods: time: sum over days
history: [2023-05-09 21:53:18] growing_degree_days: GROWING_DEGREE...
standard_name: integral_of_air_temperature_excess_wrt_time
long_name: Cumulative sum of temperature degrees for mean daily temp...
description: Annual growing degree days (mean temperature above 10 degc).Finally, xclim also allows us to call indicators using datasets and variable names.
[10]:
with xclim.set_options(cf_compliance="log"):
gdd = xclim.atmos.growing_degree_days(
tas="air", thresh="10.0 degC", freq="MS", ds=daily_ds
)
# variable names default to xclim names, so we can even do this:
renamed_daily_ds = daily_ds.rename(air="tas")
gdd = xclim.atmos.growing_degree_days(
thresh="10.0 degC", freq="MS", ds=renamed_daily_ds
)
Graphics
Xclim does not have specific functions to create graphics. However, it is built to ensure that Indices and Indicators always have appropriate axis-related metadata that libraries like `Matplotlib <https://matplotlib.org/>`__ depend on to generate detailed and informative graphics.
This graphical functionality is entirely thanks to xarray, so the following examples are applicable to generic xarray.DataArray objects. For more examples, see the directions suggested by xarray’s plotting documentation
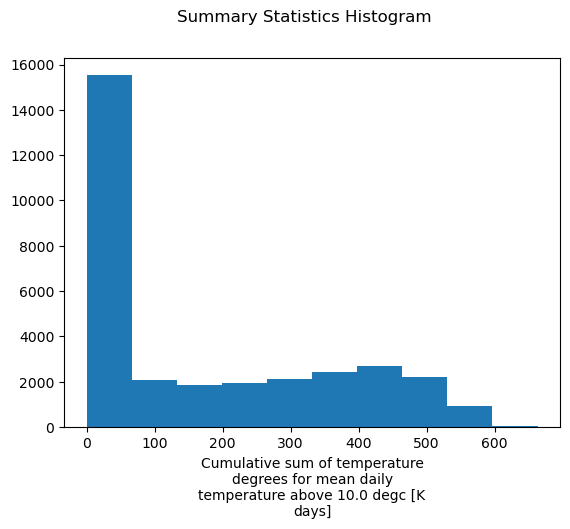
The xarray plot functions creates an histogram when the DataArray has 3 or more dimensions. In previous steps, xclim automatically filled the long_name and units attributes, which xarray uses to label the x-axis.
[11]:
import matplotlib.pyplot as plt
print("long_name:", gdd.attrs["long_name"])
print("units:", gdd.attrs["units"])
gdd.plot()
plt.suptitle("Summary Statistics Histogram")
plt.show()
long_name: Cumulative sum of temperature degrees for mean daily temperature above 10.0 degc
units: K days
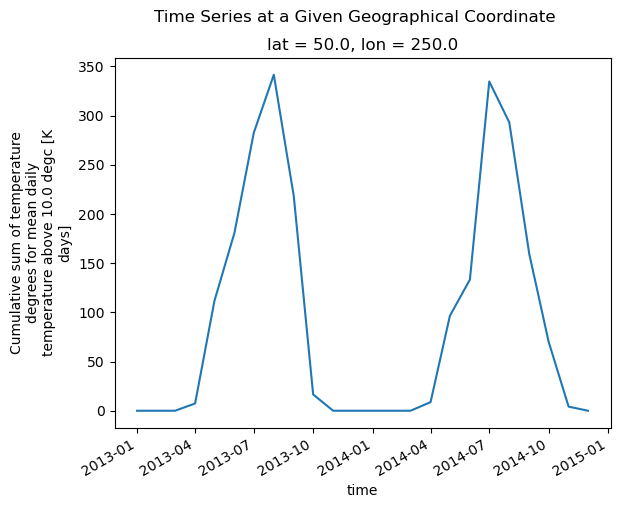
When the DataArray only has a time dimension, xarray plots a timeseries. In this case, xarray uses the long_name and units attributes provided by xclim to label the y-axis.
[12]:
gdd.isel(lon=20, lat=10).plot()
plt.suptitle("Time Series at a Given Geographical Coordinate")
plt.show()
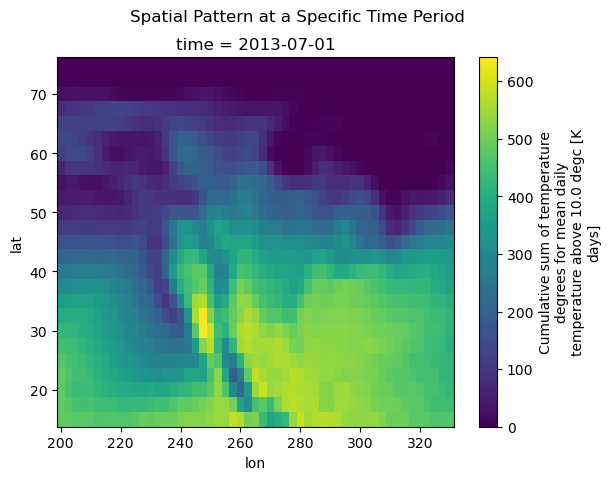
When the DataArray only has 2 dimensions, xarray plots a heatmap. In this case, xarray uses the long_name and units attributes provided by xclim to label the colorbar.
[13]:
gdd.sel(time="2013-07-01").plot()
plt.suptitle("Spatial Pattern at a Specific Time Period")
plt.show()
Writing DataArrays and Datasets to disk
To save the data as a new NetCDF, use to_netcdf:
[14]:
gdd.to_netcdf("monthly_growing_degree_days_data.nc")
It’s possible to save Dataset objects to other file formats. For more information see: xarray’s documentation